Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Filling Undefined Values#
When working with gridded data, undefined values (NaNs) can be problematic for interpolation, especially near land/sea masks. If any of the grid points used for interpolation are undefined, the result will also be undefined. This example demonstrates multiple methods to fill these undefined values and provides guidance on choosing the best method for your use case.
The Problem with Undefined Values#
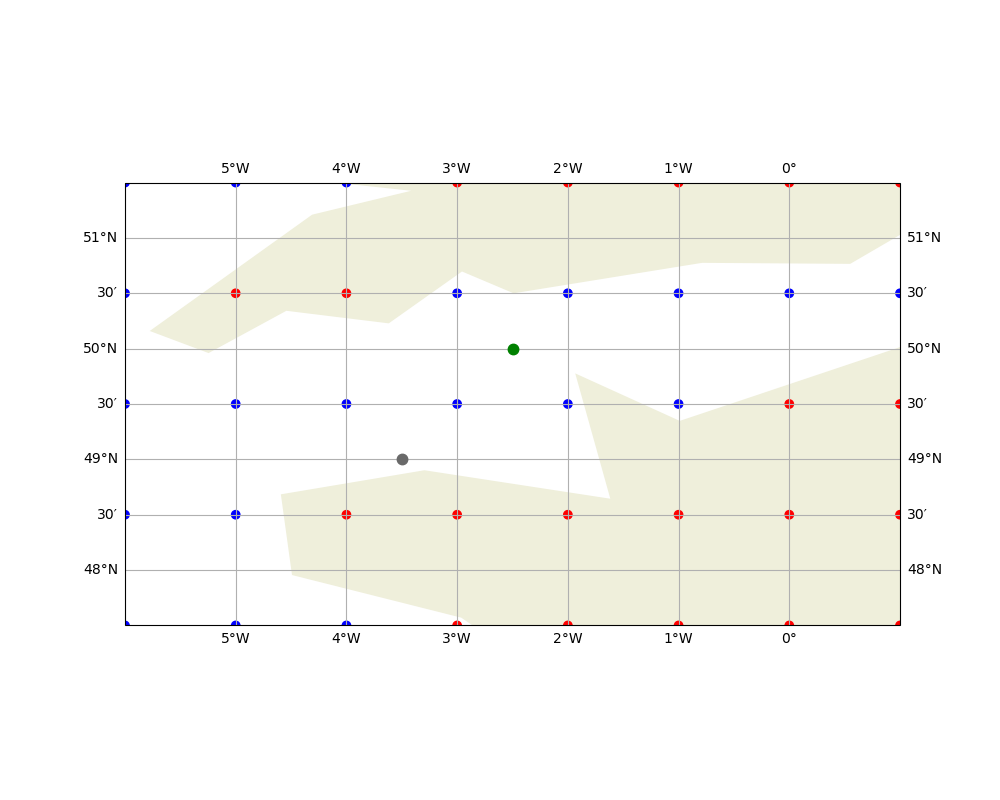
To illustrate the issue, consider the following grid where some values are undefined (represented by red points). If we want to interpolate the value at the gray point using bilinear interpolation, the calculation will fail because one of the surrounding grid points is undefined. However, the green point can be interpolated without any issues because all its surrounding points are defined.
import time
import cartopy.crs
import cartopy.feature
import matplotlib.pyplot
import numpy
import pyinterp.backends.xarray
import pyinterp.fill
import pyinterp.tests
fig = matplotlib.pyplot.figure(figsize=(10, 8))
ax = fig.add_subplot(111, projection=cartopy.crs.PlateCarree())
ax.set_extent([-6, 1, 47.5, 51.5], crs=cartopy.crs.PlateCarree())
ax.add_feature(cartopy.feature.LAND.with_scale("110m"))
ax.gridlines(draw_labels=True, dms=True, x_inline=False, y_inline=False)
lons, lats = numpy.meshgrid(
numpy.arange(-6, 2), numpy.arange(47.5, 52.5), indexing="ij"
)
mask = numpy.array(
[
[1, 1, 1, 0, 0, 0, 0, 0], # yapf: disable
[1, 1, 0, 0, 0, 0, 0, 0], # yapf: disable
[1, 1, 1, 1, 1, 1, 0, 0], # yapf: disable
[1, 0, 0, 1, 1, 1, 1, 1], # yapf: disable
[1, 1, 1, 0, 0, 0, 0, 0],
]
).T
ax.scatter(
lons.ravel(),
lats.ravel(),
c=mask,
cmap="bwr_r",
transform=cartopy.crs.PlateCarree(),
vmin=0,
vmax=1,
)
ax.plot([-3.5], [49], linestyle="", marker=".", color="dimgray", markersize=15)
ax.plot([-2.5], [50], linestyle="", marker=".", color="green", markersize=15)
fig.show()
Note
This issue does not affect nearest-neighbor interpolation, as it does not perform any arithmetic operations on the grid values.
Filling with LOESS (Local Regression)#
The pyinterp.fill.loess() function uses a fundamentally different
approach from the other methods. Instead of solving a partial differential
equation, it performs weighted local polynomial regression using a tri-cube
weight function: \(w(x)=(1-|d|^3)^3\).
When to use LOESS:
You want to smooth noisy data while filling gaps
You need local control over the filling process
The gaps are small relative to the defined regions
You want to filter or extrapolate boundary values
Limitations:
Does not guarantee smooth harmonic fills
Window size (nx, ny) must be tuned to your data
Can be slower for large grids
Let’s demonstrate with real data:
The LOESS method can fill undefined values and also filter defined ones:
filled = pyinterp.fill.loess(
grid.array,
nx=10,
ny=10,
value_type="undefined",
max_iterations=1,
is_periodic=True,
)
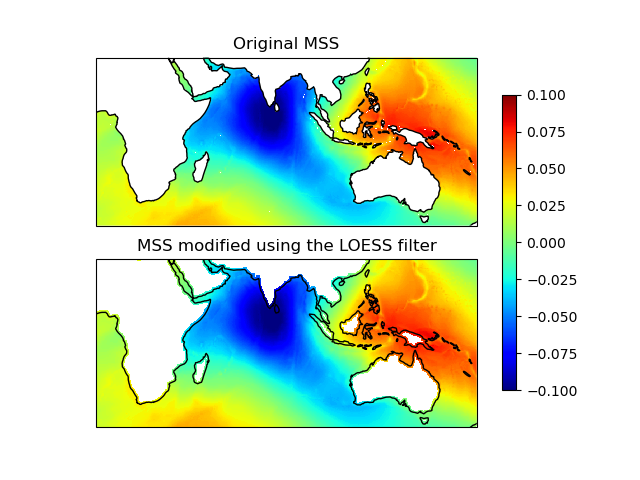
Let’s visualize the original and filled grids:
fig = matplotlib.pyplot.figure(figsize=(10, 8))
fig.subplots_adjust(left=0.05, right=0.95, top=0.95, bottom=0.05, hspace=0.25)
ax1 = fig.add_subplot(
211, projection=cartopy.crs.PlateCarree(central_longitude=180)
)
lons, lats = numpy.meshgrid(grid.x, grid.y, indexing="ij")
pcm = ax1.pcolormesh(
lons,
lats,
ds.mss.T,
cmap="jet",
shading="auto",
transform=cartopy.crs.PlateCarree(),
vmin=-0.1,
vmax=0.1,
)
ax1.coastlines()
ax1.set_title("Original MSS")
ax1.set_extent([0, 170, -45, 30], crs=cartopy.crs.PlateCarree())
ax2 = fig.add_subplot(
212, projection=cartopy.crs.PlateCarree(central_longitude=180)
)
pcm = ax2.pcolormesh(
lons,
lats,
filled,
cmap="jet",
shading="auto",
transform=cartopy.crs.PlateCarree(),
vmin=-0.1,
vmax=0.1,
)
ax2.coastlines()
ax2.set_title("Filled MSS with LOESS (nx=3, ny=3)")
ax2.set_extent([0, 170, -45, 30], crs=cartopy.crs.PlateCarree())
fig.colorbar(pcm, ax=[ax1, ax2], shrink=0.8)
<matplotlib.colorbar.Colorbar object at 0x11dc5d160>
Comparing PDE-Based Fill Methods#
The remaining methods (Gauss-Seidel, Multi-grid, and FFT Inpaint) all solve the Laplace equation to create smooth, harmonic fills. To compare them, let’s create a synthetic test case with a known analytical solution.
We’ll use a 2D Gaussian field with artificial gaps:
# Create a synthetic field
nx, ny = 200, 150
x = numpy.linspace(-5, 5, nx)
y = numpy.linspace(-5, 5, ny)
xx, yy = numpy.meshgrid(x, y, indexing="ij")
# Analytical field: 2D Gaussian
true_field = numpy.exp(-(xx**2 + yy**2) / 4)
# Create gaps (simulate land mask)
mask_gaps = numpy.zeros((nx, ny), dtype=bool)
# Central circular gap
mask_gaps[(xx**2 + yy**2) < 2] = True
# Irregular boundary gaps
mask_gaps[30:50, 40:100] = True
mask_gaps[120:140, 60:90] = True
# Apply mask
field_with_gaps = true_field.copy()
field_with_gaps[mask_gaps] = numpy.nan
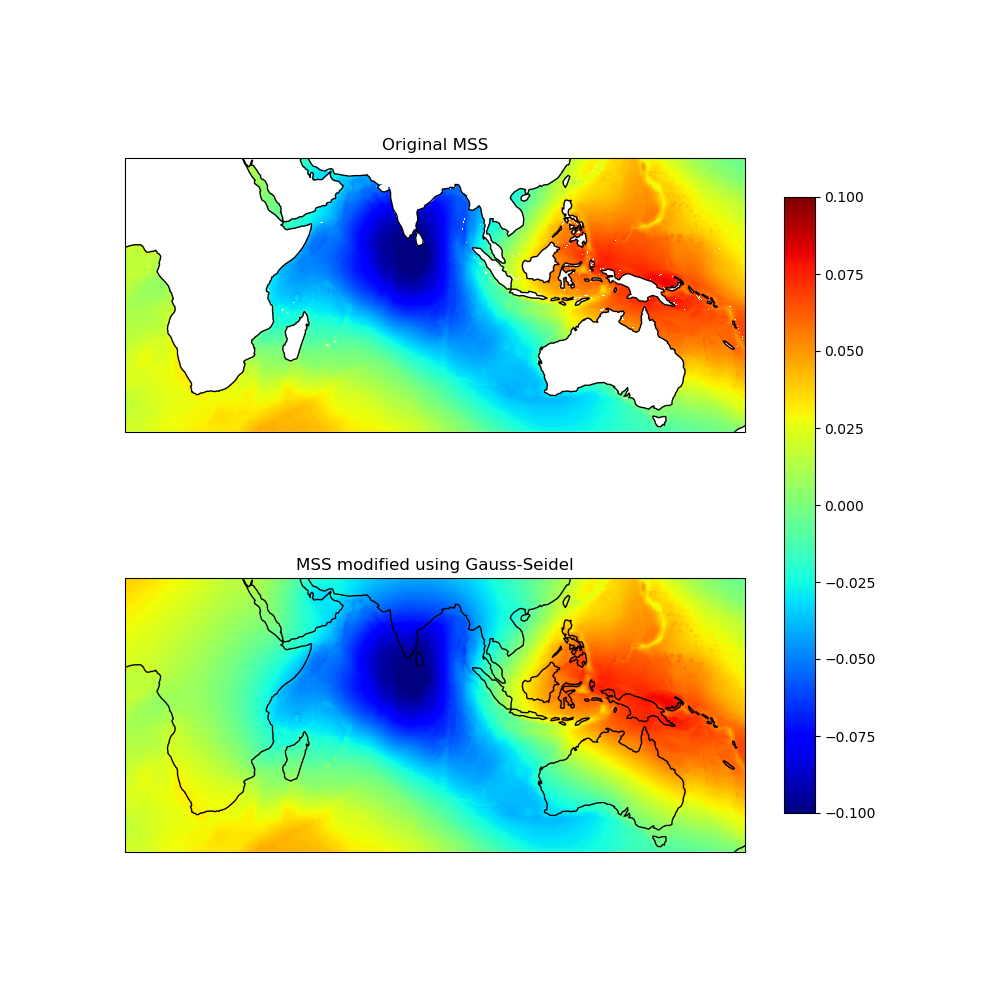
Let’s visualize the test field:
fig, axes = matplotlib.pyplot.subplots(1, 2, figsize=(12, 5))
fig.subplots_adjust(wspace=0.3)
im1 = axes[0].imshow(
true_field.T, origin="lower", cmap="viridis", aspect="auto"
)
axes[0].set_title("True Analytical Field (2D Gaussian)")
axes[0].set_xlabel("X")
axes[0].set_ylabel("Y")
fig.colorbar(im1, ax=axes[0])
im2 = axes[1].imshow(
field_with_gaps.T, origin="lower", cmap="viridis", aspect="auto"
)
axes[1].set_title("Field with Gaps (NaN values)")
axes[1].set_xlabel("X")
axes[1].set_ylabel("Y")
fig.colorbar(im2, ax=axes[1])
<matplotlib.colorbar.Colorbar object at 0x11cdd3cb0>
Gauss-Seidel Method#
pyinterp.fill.gauss_seidel() uses iterative relaxation to solve the
Laplace equation. It’s straightforward and predictable with explicit control
over relaxation parameters.
When to use Gauss-Seidel:
Small to medium-sized grids (< 500×500)
You need precise control over convergence and relaxation
Simple, predictable behavior is important
You’re debugging or validating fill behavior
filled_gs = numpy.copy(field_with_gaps)
start = time.time()
iterations_gs, residual_gs = pyinterp.fill.gauss_seidel(
filled_gs,
is_periodic=False,
epsilon=1e-4,
)
time_gs = time.time() - start
print(
f"Gauss-Seidel: {iterations_gs} iterations, "
f"residual={residual_gs:.2e}, time={time_gs:.3f}s"
)
Gauss-Seidel: 764 iterations, residual=9.96e-05, time=0.083s
Multi-grid Method#
pyinterp.fill.multi_grid() uses a V-cycle approach that solves the
problem at multiple resolution levels. It’s generally the fastest method for
large grids.
When to use Multi-grid:
Large grids (> 500×500)
You need the fastest possible solution
Convergence speed is critical
Production pipelines processing many grids
Advantages over Gauss-Seidel:
Typically 2-10× faster for large grids
Better convergence properties
More efficient memory access patterns
filled_mg = numpy.copy(field_with_gaps)
start = time.time()
iterations_mg, residual_mg = pyinterp.fill.multigrid(
filled_mg,
is_periodic=False,
epsilon=1e-4,
)
time_mg = time.time() - start
print(
f"Multi-grid: {iterations_mg} iterations, "
f"residual={residual_mg:.2e}, time={time_mg:.3f}s"
)
Multi-grid: 100 iterations, residual=1.70e-04, time=0.721s
FFT Inpaint Method#
pyinterp.fill.fft_inpaint() uses spectral methods (FFT or DCT) with
a Gaussian low-pass filter to create smooth fills.
When to use FFT Inpaint:
You want very smooth, spectrally-controlled fills
The grid has periodic or reflective boundaries
You need to control smoothness via the sigma parameter
Spectral properties of the fill matter
Trade-offs:
Can be faster for very large grids with few gaps
Requires C-contiguous arrays
The sigma parameter must be tuned
# Ensure C-contiguous array
filled_fft = numpy.ascontiguousarray(field_with_gaps.astype(numpy.float64))
start = time.time()
iterations_fft, residual_fft = pyinterp.fill.fft_inpaint(
filled_fft,
is_periodic=False,
epsilon=1e-4,
sigma=10.0,
)
time_fft = time.time() - start
print(
f"FFT Inpaint: {iterations_fft} iterations, "
f"residual={residual_fft:.2e}, time={time_fft:.3f}s"
)
FFT Inpaint: 63 iterations, residual=9.79e-05, time=0.019s
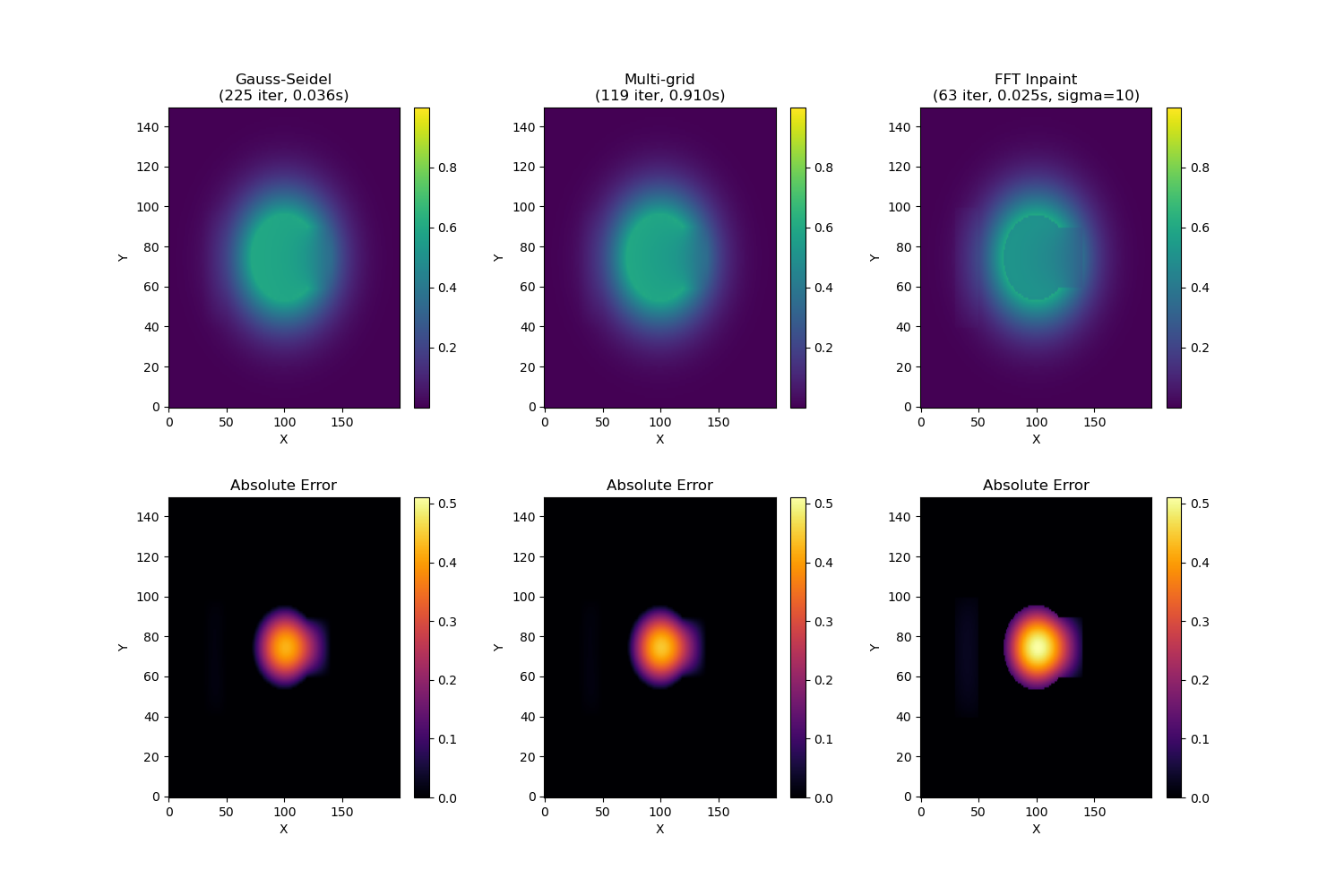
Comparing Results#
Let’s visualize all results and compute errors relative to the true field:
fig, axes = matplotlib.pyplot.subplots(2, 3, figsize=(15, 10))
fig.subplots_adjust(hspace=0.3, wspace=0.3)
# Calculate errors
error_gs = numpy.abs(filled_gs - true_field)
error_mg = numpy.abs(filled_mg - true_field)
error_fft = numpy.abs(filled_fft - true_field)
vmin, vmax = true_field.min(), true_field.max()
error_max = max(error_gs.max(), error_mg.max(), error_fft.max())
# Gauss-Seidel
im0 = axes[0, 0].imshow(
filled_gs.T,
origin="lower",
cmap="viridis",
vmin=vmin,
vmax=vmax,
aspect="auto",
)
axes[0, 0].set_title(f"Gauss-Seidel\n({iterations_gs} iter, {time_gs:.3f}s)")
axes[0, 0].set_xlabel("X")
axes[0, 0].set_ylabel("Y")
fig.colorbar(im0, ax=axes[0, 0])
im1 = axes[1, 0].imshow(
error_gs.T,
origin="lower",
cmap="inferno",
vmin=0,
vmax=error_max,
aspect="auto",
)
axes[1, 0].set_title("Absolute Error")
axes[1, 0].set_xlabel("X")
axes[1, 0].set_ylabel("Y")
fig.colorbar(im1, ax=axes[1, 0])
# Multi-grid
im2 = axes[0, 1].imshow(
filled_mg.T,
origin="lower",
cmap="viridis",
vmin=vmin,
vmax=vmax,
aspect="auto",
)
axes[0, 1].set_title(f"Multi-grid\n({iterations_mg} iter, {time_mg:.3f}s)")
axes[0, 1].set_xlabel("X")
axes[0, 1].set_ylabel("Y")
fig.colorbar(im2, ax=axes[0, 1])
im3 = axes[1, 1].imshow(
error_mg.T,
origin="lower",
cmap="inferno",
vmin=0,
vmax=error_max,
aspect="auto",
)
axes[1, 1].set_title("Absolute Error")
axes[1, 1].set_xlabel("X")
axes[1, 1].set_ylabel("Y")
fig.colorbar(im3, ax=axes[1, 1])
# FFT Inpaint
im4 = axes[0, 2].imshow(
filled_fft.T,
origin="lower",
cmap="viridis",
vmin=vmin,
vmax=vmax,
aspect="auto",
)
axes[0, 2].set_title(
f"FFT Inpaint\n({iterations_fft} iter, {time_fft:.3f}s, sigma=10)"
)
axes[0, 2].set_xlabel("X")
axes[0, 2].set_ylabel("Y")
fig.colorbar(im4, ax=axes[0, 2])
im5 = axes[1, 2].imshow(
error_fft.T,
origin="lower",
cmap="inferno",
vmin=0,
vmax=error_max,
aspect="auto",
)
axes[1, 2].set_title("Absolute Error")
axes[1, 2].set_xlabel("X")
axes[1, 2].set_ylabel("Y")
fig.colorbar(im5, ax=axes[1, 2])
fig.show()
Summary#
Gauss-Seidel: Slowest but simplest. Good for small grids or when you need fine-grained control.
Multi-grid: Fastest for large grids. The recommended default for most use cases.
FFT Inpaint: Excellent for creating spectrally smooth fills. Requires tuning of the sigma parameter.
LOESS: A different approach suitable for noisy data or when local smoothing is desired.
Choose the method that best fits your grid size, performance needs, and desired fill characteristics.
Total running time of the script: (0 minutes 4.480 seconds)