Note
Go to the end to download the full example code or to run this example in your browser via Binder.
3D Interpolation#
This example demonstrates how to perform 3D interpolation on a regular grid. The pyinterp library supports both trivariate and bicubic interpolation for 3D data.
import cartopy.crs
import matplotlib
import matplotlib.pyplot
import numpy
import pyinterp
import pyinterp.backends.xarray
import pyinterp.tests
Trivariate Interpolation#
Trivariate interpolation builds upon bivariate interpolation by adding a
third dimension. It first performs bilinear interpolation on the 2D spatial
plane (longitude and latitude), followed by linear interpolation along the
time dimension. Alternatively, you can opt for nearest-neighbor interpolation
on the time axis by specifying third_axis='nearest'. First, we load the
3D dataset and create the interpolator object.
Next, we define the coordinates for interpolation. To avoid interpolating at the exact grid points, we introduce a slight shift.
mx, my, mz = numpy.meshgrid(
numpy.arange(-180, 180, 0.25) + 1 / 3.0,
numpy.arange(-80, 80, 0.25) + 1 / 3.0,
numpy.array(["2002-07-02T15:00:00"], dtype="datetime64"),
indexing="ij",
)
Now, we perform the trivariate interpolation.
trivariate = interpolator.trivariate(
{"longitude": mx.ravel(), "latitude": my.ravel(), "time": mz.ravel()}
)
Windowed Interpolators#
Bicubic and spline interpolation provides smoother results compared to
bilinear by using a windowed approach that considers a neighborhood of grid
points within a calculation window. You can use it by passing
method='bicubic' to the trivariate method or other supported methods.
interpolator = pyinterp.backends.xarray.Grid3D(ds.data_vars["tcw"])
We then perform the bicubic interpolation.
bicubic = interpolator.trivariate(
{"longitude": mx.ravel(), "latitude": my.ravel(), "time": mz.ravel()},
method="bicubic",
half_window_size_x=3,
half_window_size_y=3,
boundary_mode="shrink",
)
To visualize the results, we reshape the output arrays and extract the longitude and latitude coordinates.
trivariate = trivariate.reshape(mx.shape).squeeze(axis=2)
bicubic = bicubic.reshape(mx.shape).squeeze(axis=2)
lons = mx[:, 0].squeeze()
lats = my[0, :].squeeze()
Finally, let’s plot the results of both trivariate and bicubic interpolation.
fig = matplotlib.pyplot.figure(figsize=(10, 8))
fig.subplots_adjust(left=0.05, right=0.95, top=0.95, bottom=0.05, hspace=0.25)
ax1 = fig.add_subplot(
211, projection=cartopy.crs.PlateCarree(central_longitude=180)
)
ax1.set_extent([-180, 180, -90, 90], crs=cartopy.crs.PlateCarree())
pcm = ax1.pcolormesh(
lons,
lats,
trivariate.T,
cmap="jet",
shading="auto",
transform=cartopy.crs.PlateCarree(),
)
ax1.coastlines()
ax1.set_title("Trivariate Interpolation")
ax2 = fig.add_subplot(
212, projection=cartopy.crs.PlateCarree(central_longitude=180)
)
ax2.set_extent([-180, 180, -90, 90], crs=cartopy.crs.PlateCarree())
pcm = ax2.pcolormesh(
lons,
lats,
bicubic.T,
cmap="jet",
shading="auto",
transform=cartopy.crs.PlateCarree(),
)
ax2.coastlines()
ax2.set_title("Bicubic Interpolation")
fig.colorbar(pcm, ax=[ax1, ax2], shrink=0.8)
<matplotlib.colorbar.Colorbar object at 0x11df5cc20>
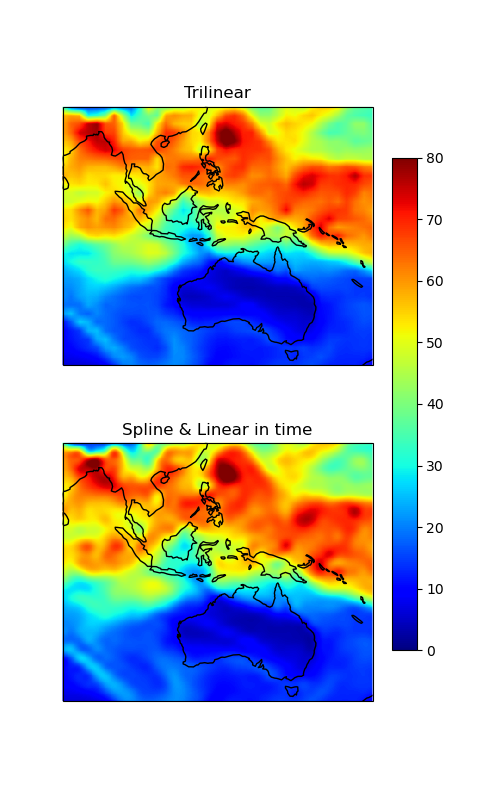
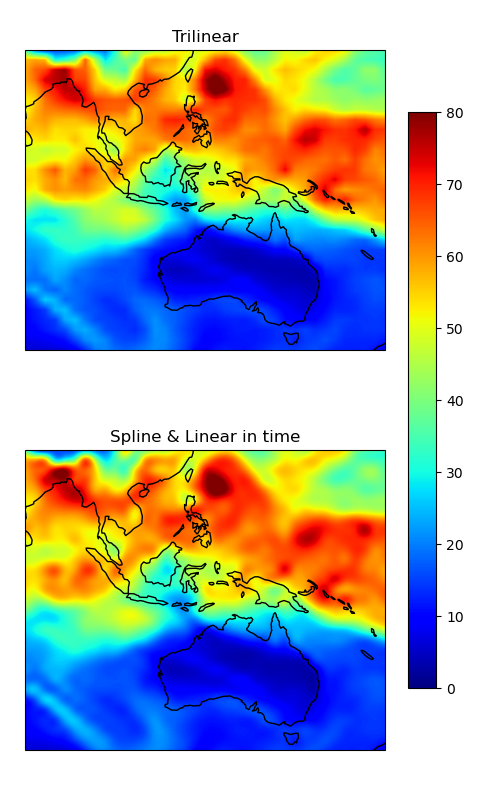
Same plot as above, but zoomed into a specific region to better highlight the differences between the two interpolation methods.
fig = matplotlib.pyplot.figure(figsize=(5, 8))
fig.subplots_adjust(left=0.05, right=0.95, top=0.95, bottom=0.05, hspace=0.25)
ax1 = fig.add_subplot(
211, projection=cartopy.crs.PlateCarree(central_longitude=180)
)
pcm = ax1.pcolormesh(
lons,
lats,
trivariate.T,
cmap="jet",
shading="auto",
transform=cartopy.crs.PlateCarree(),
vmin=0,
vmax=80,
)
ax1.coastlines()
ax1.set_extent([80, 170, -45, 30], crs=cartopy.crs.PlateCarree())
ax1.set_title("Trilinear")
ax2 = fig.add_subplot(
212, projection=cartopy.crs.PlateCarree(central_longitude=180)
)
pcm = ax2.pcolormesh(
lons,
lats,
bicubic.T,
cmap="jet",
shading="auto",
transform=cartopy.crs.PlateCarree(),
vmin=0,
vmax=80,
)
ax2.coastlines()
ax2.set_extent([80, 170, -45, 30], crs=cartopy.crs.PlateCarree())
ax2.set_title("Spline & Linear in time")
fig.colorbar(pcm, ax=[ax1, ax2], shrink=0.8)
<matplotlib.colorbar.Colorbar object at 0x11cd056a0>
Total running time of the script: (0 minutes 6.956 seconds)