map_of_fle#
The map_of_fle script computes global or regional maps of Finite-Size (FSLE)
or Finite-Time (FTLE) Lyapunov Exponents from a time series of velocity fields
stored in NetCDF files.
Usage#
usage: map_of_fle [--resolution STEP] [--x_min X_MIN] [--x_max X_MAX]
[--y_min Y_MIN] [--y_max Y_MAX] [--mode {fsle,ftle}]
[--time_direction {backward,forward}]
[--stencil {triplet,quintuplet}]
[--initial_separation DELTA] [--advection_time DURATION]
[--final_separation DELTA]
[--integration_time_step DURATION] [--diagnostic]
[--threads THREADS] [--unit {metric,angular}]
[--mask PATH VARNAME] [--verbose] [--version] [--help]
configuration output start_time
Compute Map of FLE
positional arguments:
configuration configuration file
output map of FLE in NetCDF format
start_time start of the integration
options:
--verbose Verbose mode
--version print the version number and exit
--help, -h show this help message and exit
grid arguments:
Set the grid properties to create
--resolution STEP output grid resolution in degrees
--x_min X_MIN x min in degrees
--x_max X_MAX x max in degrees
--y_min Y_MIN y min in degrees
--y_max Y_MAX y max in degrees
integration arguments:
Define integration
--mode {fsle,ftle} define the integration mode: fsle to compute Finite
Size Lyapunov Exponent, ftle to compute Finite Time
Lyapunov Exponent.
--time_direction {backward,forward}
time integration direction
--stencil {triplet,quintuplet}
type of stencil used to compute the finite difference.
--initial_separation DELTA
initial separation in degrees of neighbouring
particules
--advection_time DURATION
time of advection in number of days elapsed since the
beginning of the simulation. This option set the
maximum time of advection in FSLE mode
--final_separation DELTA
maximum final separation in degrees
--integration_time_step DURATION
particles time integration step in hours
--diagnostic write additional variables containing diagnostics,
depending on calculation mode. These variables are the
effective final separation distance and advection
time, in FSLE mode, or the effective final separation
distance, in FTLE mode.
--threads THREADS number of threads to use for the computation. If 0 all
CPUs are used. If 1 is given, no parallel computing
code is used at all.
reader arguments:
Set options of the NetCDF reader.
--unit {metric,angular}
system of units for velocity
--mask PATH VARNAME netCDF grid for fixing undefined cells
Summary#
Input: a list of NetCDF files providing eastward (U) and northward (V) velocities on a lon/lat grid over time.
Output: a NetCDF file with variables
lambda1,lambda2(FLE/FTLE) andtheta1,theta2(eigenvector orientations), plus optional diagnostics.Modes:
fsle(default) orftlevia--mode.
Quick start#
Example: FSLE map over a subdomain, forward in time, 0.05° resolution, 89 days of advection and 0.2° final separation:
map_of_fle list.ini fsle.nc "2010-01-01" \
--mode fsle --time_direction forward --advection_time 89 \
--resolution 0.05 --x_min 40 --x_max 60 --y_min -60 --y_max -40 \
--final_separation 0.2 --verbose
The output NetCDF will contain variables described in “Output” below.
Inputs#
Data description#
The files required to run this example are located in the data.zip archive. These files describe a time series of eastward and northward velocities on a global grid. You can download up-to-date files from the AVISO website.
Each file describes a single time step. The NetCDF header typically contains variables similar to:
dimensions:
y = 915 ;
x = 1080 ;
variables:
double y(y) ;
y:long_name = "Latitudes" ;
y:units = "degrees_north" ;
double x(x) ;
x:long_name = "Longitudes" ;
x:units = "degrees_east" ;
float u(y, x) ;
u:_FillValue = 999.f ;
u:long_name = "eastward velocity" ;
u:units = "cm/s" ;
float v(y, x) ;
v:_FillValue = 999.f ;
v:long_name = "northward velocity" ;
v:units = "cm/s" ;
Note: Units like cm/s are metric. Keep --unit metric (default) for such
inputs. Use --unit angular only if your U/V are provided in angular units
(e.g., degrees per time).
Configuration file#
You must provide a simple .ini-like configuration file listing the U and V
files and variable names. Example:
U = /path/to/dt_upd_global_merged_madt_uv_20091230_20091230_20110329.nc
U = /path/to/dt_upd_global_merged_madt_uv_20100106_20100106_20110329.nc
... (one line per time slice)
V = /path/to/dt_upd_global_merged_madt_uv_20091230_20091230_20110329.nc
V = /path/to/dt_upd_global_merged_madt_uv_20100106_20100106_20110329.nc
... (one line per time slice)
U_NAME = Grid_0001
V_NAME = Grid_0002
FILL_VALUE = 0
U: paths to NetCDF files containing eastward velocities (one per line).V: paths to NetCDF files containing northward velocities (one per line).U_NAME/V_NAME: variable names inside each NetCDF file for U and V.FILL_VALUE: value to use when encountering missing data. Use0to avoid propagation of missing values, ornanif you want missing values to propagate through the computation.
Paths can be absolute or use environment variables, e.g.:
U = ${DATA}/dt_upd_global_merged_madt_uv_20100407_20100407_20110329.nc
Options and modes#
Common options#
--resolution: output grid step in degrees (default: 1.0). Also used as default--initial_separationif not provided.--x_min/x_max/y_min/y_max: lon/lat bounds of the output grid.--integration_time_step: particle integration time step in hours (default: 6).--unit: velocity unit system (metricorangular), defaultmetric. Choose according to U/V variable units in your files.--mask PATH VARNAME: a NetCDF grid; cells that are masked in this grid are skipped to speed up computation.--threads N: number of CPU threads.0uses all CPUs;1disables multithreading.--verbose: print debug information.
FSLE vs FTLE#
--mode fsle(default): FSLE computation requires a--final_separation(in degrees).--advection_timesets the maximum advection time.--mode ftle: FTLE computation ignores--final_separationand uses the exact--advection_time.--time_direction:forwardorbackwardintegration.
Constraints#
--final_separationis not allowed in FTLE mode and will raise an error.--initial_separationdefaults to--resolutionwhen unspecified.
Distributed execution (Dask)#
You can split the computation by latitude chunks and distribute it with Dask:
--local-cluster: start a local Dask cluster automatically (for testing or single-node workflows).--scheduler_file PATH: connect to an existing Dask cluster using a scheduler file.
Examples:
# Local cluster
map_of_fle list.ini fsle.nc "2010-01-01" --mode fsle \
--time_direction forward --advection_time 89 --final_separation 0.2 \
--resolution 0.05 --x_min 40 --x_max 60 --y_min -60 --y_max -40 \
--local-cluster --verbose
# External cluster
map_of_fle list.ini fsle.nc "2010-01-01" --mode fsle \
--time_direction forward --advection_time 89 --final_separation 0.2 \
--resolution 0.05 --x_min 40 --x_max 60 --y_min -60 --y_max -40 \
--scheduler_file /path/to/scheduler.json
Output#
The output NetCDF contains:
Dimensions:
lon,lat(note the variable dimensionality is(lon, lat)).Coordinates: -
lon[degrees_east] -lat[degrees_north]Main variables:
theta1[degree]: orientation of eigenvectors associated with maximum eigenvalues of the Cauchy–Green strain tensor.theta2[degree]: orientation of eigenvectors associated with minimum eigenvalues of the Cauchy–Green strain tensor.lambda1[1/day]: FLE/FTLE associated with maximum eigenvalues.lambda2[1/day]: FLE/FTLE associated with minimum eigenvalues.
Optional diagnostics (
--diagnostic):separation_distance[degree]: effective final separation distance.advection_time[day]: actual advection time (FSLE mode only), counted since the provided start time.
Examples#
FSLE forward#
map_of_fle.py list.ini fsle.nc "2010-01-01" --advection_time 89 \
--resolution 0.05 --x_min 40 --x_max 60 --y_min -60 --y_max -40 \
--final_separation 0.2 --verbose --time_direction forward
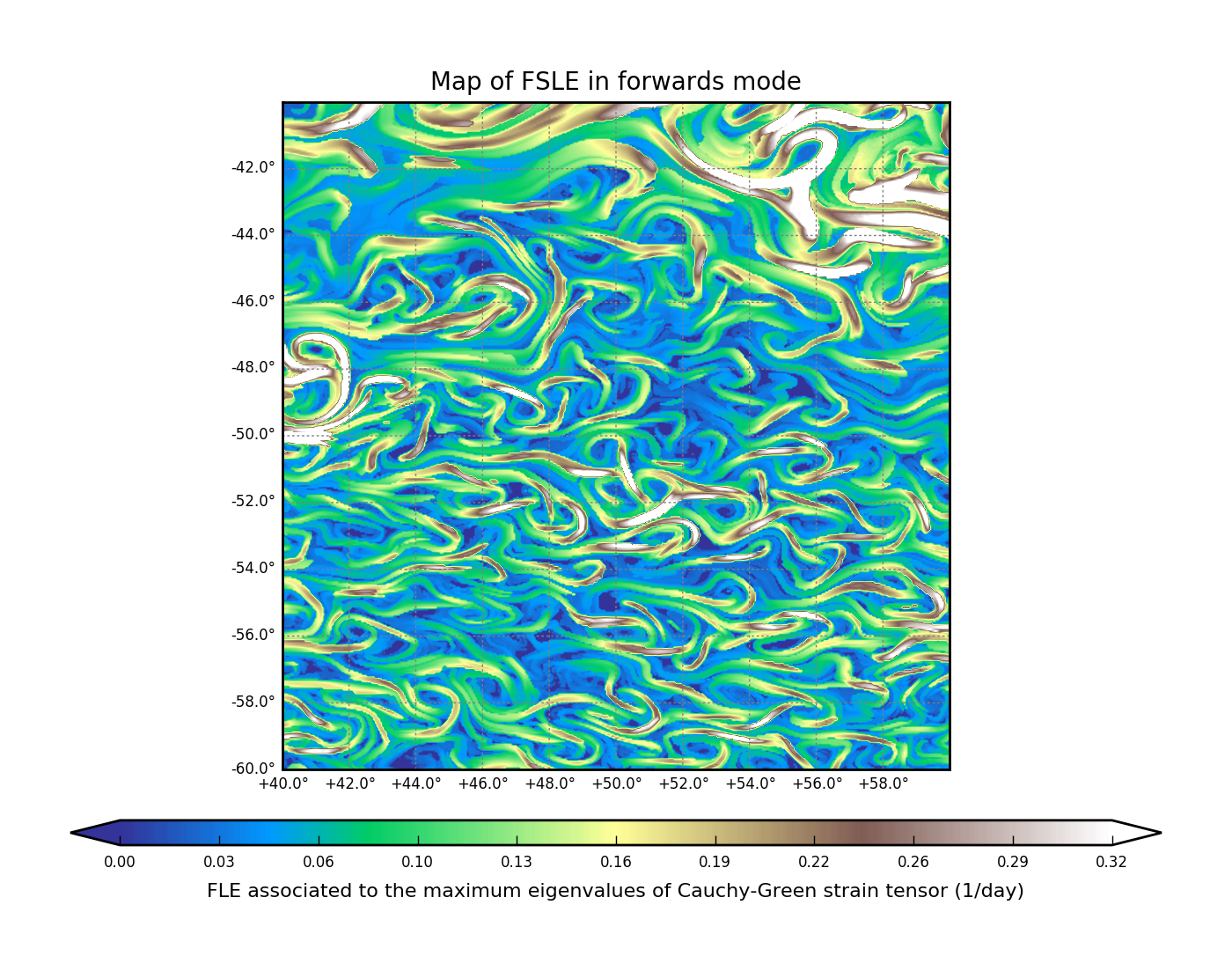
FSLE backward#
map_of_fle.py list.ini fsle_backwards.nc "2010-03-31" --advection_time 89 \
--resolution 0.05 --x_min 40 --x_max 60 --y_min -60 --y_max -40 \
--final_separation 0.2 --verbose --time_direction backward
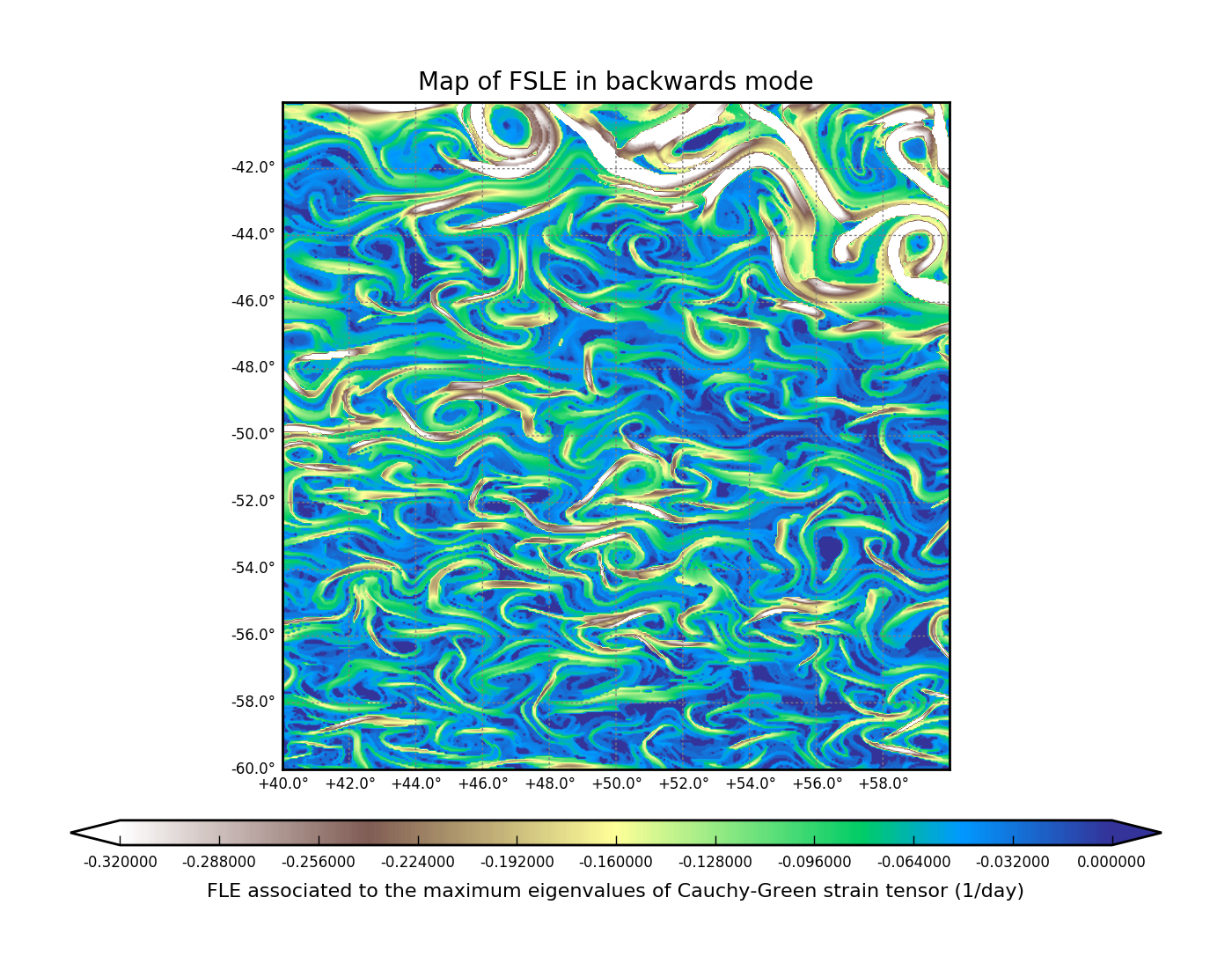
FTLE example#
map_of_fle list.ini ftle.nc "2010-01-01" --mode ftle --advection_time 60 \
--resolution 0.05 --x_min 40 --x_max 60 --y_min -60 --y_max -40 \
--verbose --time_direction forward
Masking land/invalid cells#
Use a land/sea mask (or any boolean mask) to skip cells and speed up run time:
map_of_fle list.ini fsle.nc "2010-01-01" --advection_time 89 \
--final_separation 0.2 --mask mask.nc land
Help#
Type map_of_fle --help or map_of_fle.py --help to see all options.
Troubleshooting#
PYTHONPATH#
If you see this error message:
Traceback (most recent call last):
File "map_of_fle.py", line 17, in <module> import lagrangian
ImportError: No module named lagrangian
Set the PYTHONPATH environment variable to include the directory that
contains the compiled lagrangian module (lagrangian.so).
UDUNITS2_XML_PATH#
If you see this error message:
RuntimeError: The variable UDUNITS2_XML_PATH is unset, and the installed, default unit, database couldn't be opened: No such file or directory
Set the UDUNITS2 database path, for example:
export UDUNITS2_XML_PATH=/path/to/share/udunits/udunits2.xml
Dask cluster connection#
If
--local-clusterhangs with zero workers, ensure thedaskanddistributedpackages are installed in your environment.When using
--scheduler_file, verify that the file is readable and points to a reachable scheduler.
See also#
metric_to_angular: convert metric velocity fields to angular units when needed.
path: compute particle trajectories using the same time series inputs.